Sample name: gold4
Looper stats summary
| sample_name |
gold4 |
| sample_description |
ATAC-seq from dendritic cell (ENCLB586KIS) |
| treatment_description |
Homo sapiens dendritic in vitro differentiated cells trea... |
| organism |
human |
| protocol |
ATAC-seq |
| data_source |
SRA |
| SRR |
SRR5210428 |
| SRX |
SRX2523884 |
| Sample_geo_accession |
GSM2471269 |
| Sample_series_id |
GSE94196 |
| read_type |
PAIRED |
| Sample_instrument_model |
Illumina HiSeq 2000 |
| read1 |
/project/shefflab/data/sra_fastq//SRR5210428_1.fastq.gz |
| read2 |
/project/shefflab/data/sra_fastq//SRR5210428_2.fastq.gz |
| Raw_reads |
101078490 |
| Fastq_reads |
101078490 |
| Trimmed_reads |
101077540 |
| Trim_loss_rate |
0.0 |
| Mapped_reads |
78149573 |
| QC_filtered_reads |
6702363 |
| Aligned_reads |
71447210 |
| Alignment_rate |
70.69 |
| Total_efficiency |
70.68 |
| Mitochondrial_reads |
6339 |
| NRF |
0.94 |
| PBC1 |
0.97 |
| PBC2 |
35.97 |
| Unmapped_reads |
1368463 |
| Duplicate_reads |
1038863 |
| Dedup_aligned_reads |
70408347.0 |
| Dedup_alignment_rate |
69.66 |
| Dedup_total_efficiency |
69.66 |
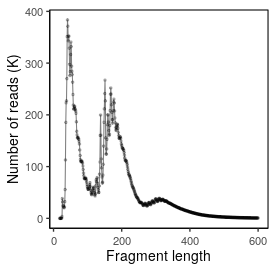
| Read_length |
42 |
| Genome_size |
3099922541 |
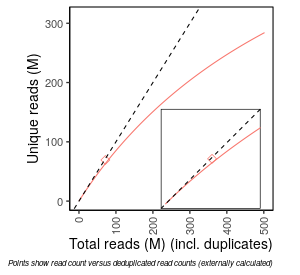
| Frac_exp_unique_at_10M |
0.9785 |
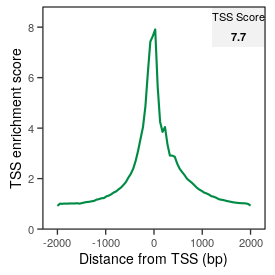
| TSS_score |
7.7 |
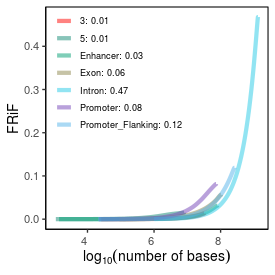
| Peak_count |
305045 |
| File_mb |
3136 |
| Read_type |
PAIRED |
| Genome |
hg38 |
| Aligned_reads_rCRSd |
14045392.0 |
| Alignment_rate_rCRSd |
13.9 |
| Aligned_reads_human_repeats |
7514112.0 |
| Alignment_rate_human_repeats |
7.43 |
| Time |
0:11:41 |
| Success |
09-15-12:29:13 |