Looper PEPATAC_tutorial summary
| sample_name |
protocol |
organism |
read1 |
read2 |
read_type |
File_mb |
Read_type |
Genome |
Raw_reads |
Fastq_reads |
Trimmed_reads |
Trim_loss_rate |
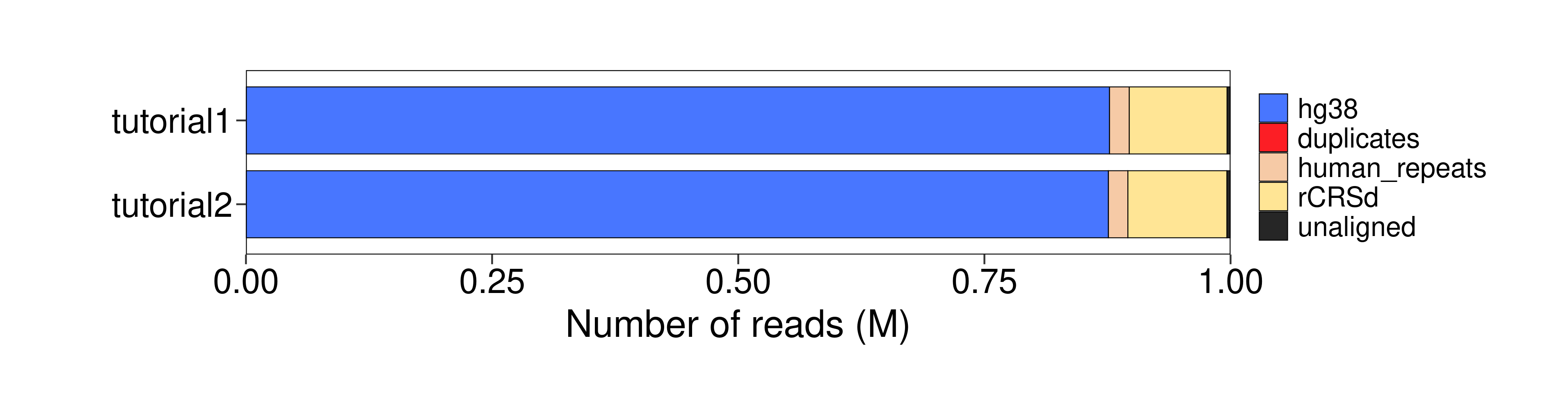
Aligned_reads_rCRSd |
Alignment_rate_rCRSd |
Aligned_reads_human_repeats |
Alignment_rate_human_repeats |
Mapped_reads |
QC_filtered_reads |
Aligned_reads |
Alignment_rate |
Total_efficiency |
Mitochondrial_reads |
NRF |
PBC1 |
PBC2 |
Unmapped_reads |
Duplicate_reads |
Dedup_aligned_reads |
Dedup_alignment_rate |
Dedup_total_efficiency |
Read_length |
Genome_size |
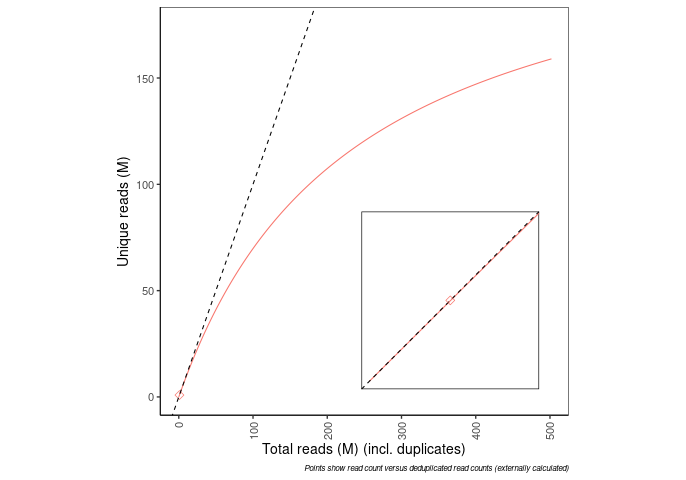
Frac_exp_unique_at_10M |
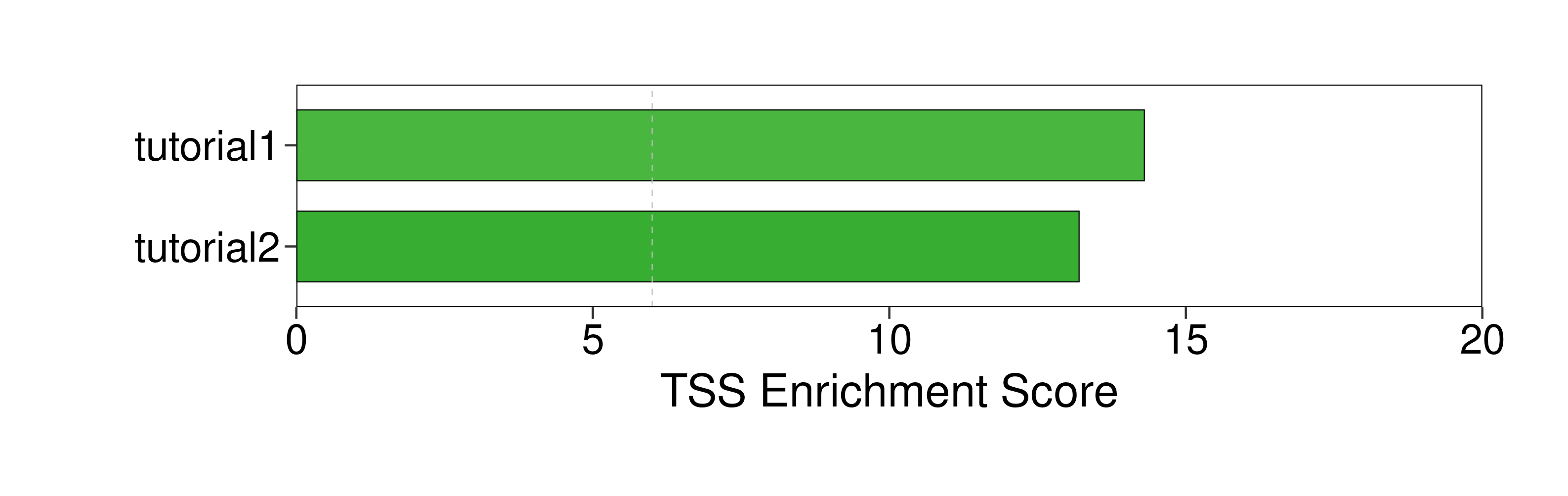
TSS_score |
Peak_count |
FRiP |
Time |
Success |
|
tutorial1
|
ATAC |
human |
/scratch/jps3dp/data/tmp/pepatac_tutorial/tools/pepatac/e... |
/scratch/jps3dp/data/tmp/pepatac_tutorial/tools/pepatac/e... |
paired |
27 |
paired |
hg38 |
1000000 |
1000000 |
1000000 |
0.0 |
99360.0 |
9.94 |
20102.0 |
2.01 |
880479 |
3539 |
876940 |
87.69 |
87.69 |
18 |
1.0 |
1.0 |
438465.0 |
59 |
0 |
876940.0 |
87.69 |
87.69 |
42 |
3099922541 |
0.9572 |
14.3 |
535285 |
0.92 |
0:38:48 |
09-21-14:43:02 |
|
tutorial2
|
ATAC |
human |
/scratch/jps3dp/data/tmp/pepatac_tutorial/tools/pepatac/e... |
/scratch/jps3dp/data/tmp/pepatac_tutorial/tools/pepatac/e... |
paired |
27 |
paired |
hg38 |
1000000 |
1000000 |
1000000 |
0.0 |
100556.0 |
10.06 |
19842.0 |
1.98 |
879537 |
3707 |
875830 |
87.58 |
87.58 |
30 |
1.0 |
1.0 |
437908.0 |
65 |
0 |
875830.0 |
87.58 |
87.58 |
42 |
3099922541 |
NA |
13.2 |
534448 |
0.92 |
0:39:02 |
09-21-15:22:06 |